Using datapasta to create R objects from cut-and-paste data
Being able to paste data into source code documents is useful for all sorts of reasons, not least because it allows for a reproducible example, also known as a reprex—a minimal, self-containedexample that demonstrates a problem or a concept. By including data in the source code, others can run the code and see the results for themselves, making it easier to understand and replicate the results.
The R datapasta package makes it easy to paste data into R source code documents. It provides a set of functions for converting data to and from R definitions and is extremely useful when creating static data objects in code examples, tests, or when sharing. In this recipe, you will learn how to use datapasta to bring external data into your source code documents by typing them in longhand.
Getting ready
We will use the datapasta package, though installing it is non-standard. Use this command:
renv::install("datapasta", repos = c(mm = "https://milesmcbain.r-universe.dev", getOption("repos"))) This should install the package using renv and make us ready to go. Remember to install renv the usual way if you don’t already have it.
How to do it…
The datapasta tool is implemented as an add-in for RStudio, so we begin by setting that up:
- Use the RStudio Tools | Addins | Browse Addins menu and then Keyboard Shortcuts. You get to choose which key combination you want to use for pasting.
- Click the middle column next to the operations, as shown in the following screenshot, and press the keys you want to use. The combination in Figure 2.1 is a good choice:
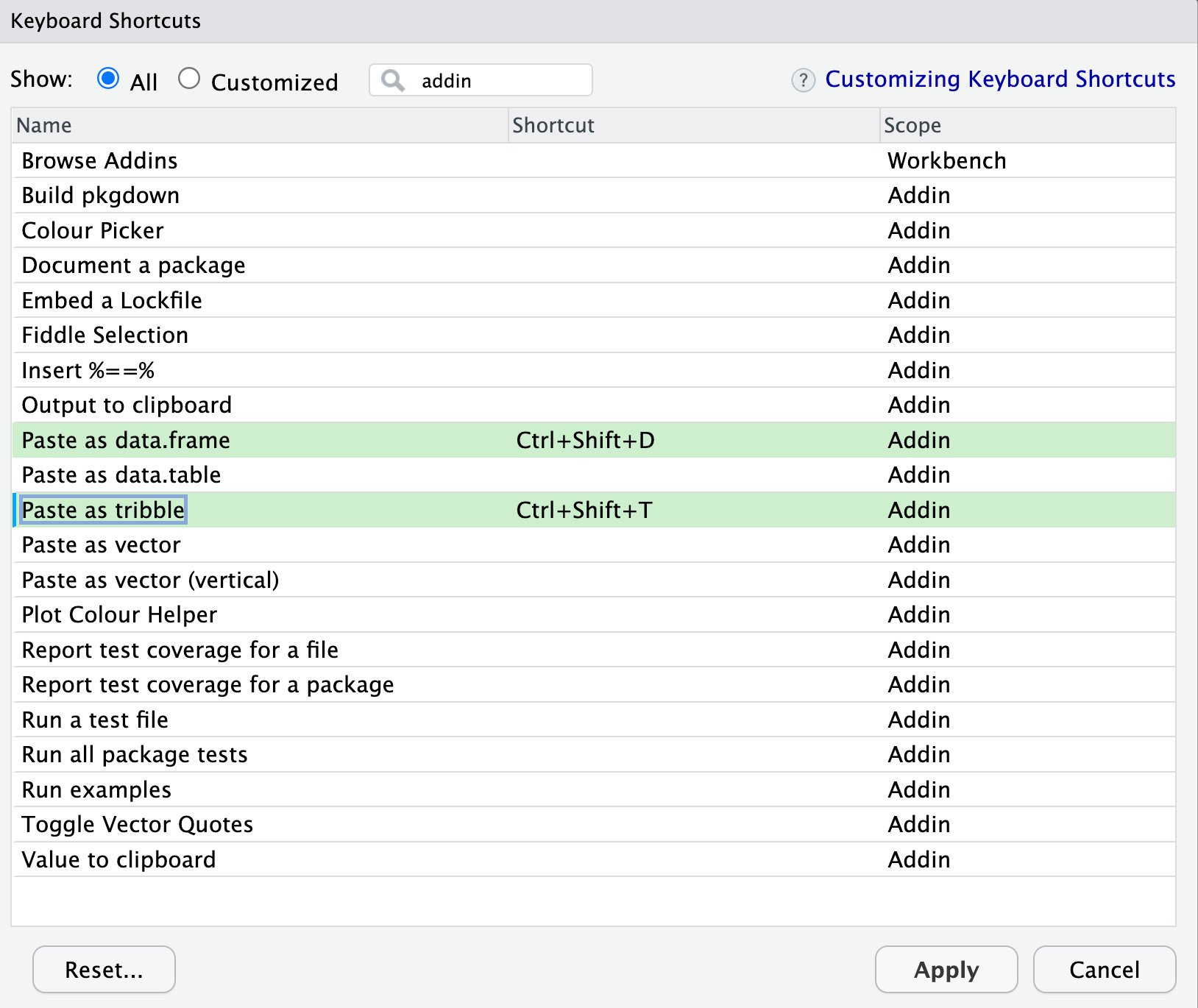
Figure 2.1 – Selecting key shortcuts
- Get a web table—for example, go to this page on Wikipedia: https://en.wikipedia.org/wiki/Tab-separated_values—and copy the whole text-based example table using the browser’s right-click Copy feature. This should put the text table in your copy/paste buffer. It should look like what’s shown in Figure 2.2:
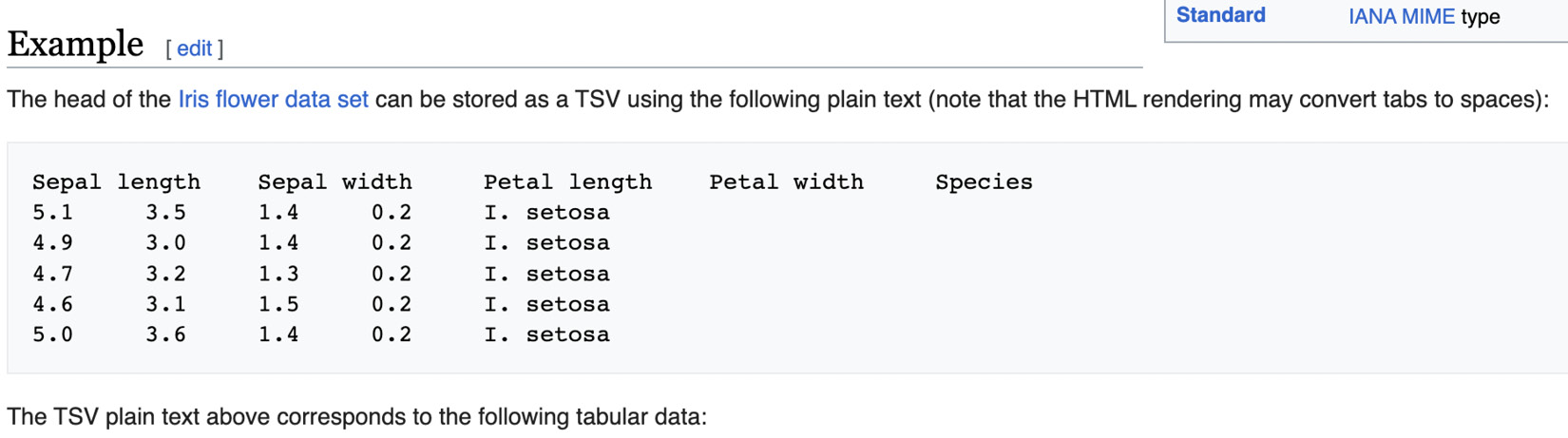
Figure 2.2 – Web data on Wikipedia
- Paste the table now in your copy/paste buffer into an R source document. Place the typing cursor at a suitable place in the source R document you’re working in and use the key combo to paste in the table.
So, with the preceding setup, we should be able to quickly take data from varied sources and coerce them into R objects for analysis.
How it works…
The datapasta package is really useful. The first step sets up our preferred paste keys for later use; the second step is simple, and we just go somewhere and find some data in the world we would like in our R source; and by the third step, we’re selecting the place to put the data definition and pasting it in. Our example goes from a table in a web page to this definition for an R object:
data.frame( stringsAsFactors = FALSE,
Sepal.length = c(5.1, 4.9, 4.7, 4.6, 5),
Sepal.width = c(3.5, 3, 3.2, 3.1, 3.6),
Petal.length = c(1.4, 1.4, 1.3, 1.5, 1.4),
Petal.width = c(0.2, 0.2, 0.2, 0.2, 0.2),
Species = c("I. setosa","I. setosa",
"I. setosa","I. setosa","I. setosa")
)
And that powerful little operation is how we can convert web data to source code very easily.
There’s more…
It’s possible to go the other way around, from some R object to a definition of that object using the dpasta() function, which can coerce data frames, tibbles, and vectors into definitions. This is really useful for reproducible examples. This example shows how to actually build the object with code so that we don’t have to share the file and everything is in one source document:
library(datapasta)mtcars |> dpasta()
This shows how the datapasta package is a super-useful tool for turning objects from the web and within R into code.

