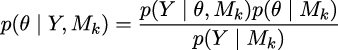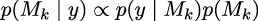Chapter 5
Comparing Models
A map is not the territory it represents, but, if correct, it has a similar structure to the territory. – Alfred Korzybski
Models should be designed as approximations to help us understand a particular problem or a class of related problems. Models are not designed to be verbatim copies of the real world. Thus, all models are wrong in the same sense that maps are not the territory. But not all models are equally wrong; some models will be better than others at describing a given problem.
In the previous chapters, we focused our attention on the inference problem, that is, how to learn the values of parameters from data. In this chapter, we are going to focus on a complementary problem: how to compare two or more models for the same data. As we will learn, this is both a central problem in data analysis and a tricky one. In this chapter, we are going to keep examples super simple, so we can focus on the technical aspects of model comparison. In...