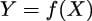Chapter 8
Gaussian Processes
Lonely? You have yourself. Your infinite selves. - Rick Sanchez (at least the one from dimension C-137)
In the last chapter, we learned about the Dirichlet process, an infinite-dimensional generalization of the Dirichlet distribution that can be used to set a prior on an unknown continuous distribution. In this chapter, we will learn about the Gaussian process, an infinite-dimensional generalization of the Gaussian distribution that can be used to set a prior on unknown functions. Both the Dirichlet process and the Gaussian process are used in Bayesian statistics to build flexible models where the number of parameters is allowed to increase with the size of the data.
We will cover the following topics:
Functions as probabilistic objects
Kernels
Gaussian processes with Gaussian likelihoods
Gaussian processes with non-Gaussian likelihoods
Hilbert space Gaussian process


 the inverse link function and
the inverse link function and