In this section, we will briefly demonstrate the usefulness of Jupyter Notebooks with examples. Then, we'll walk through the basics of how they work and how to run them within the Jupyter platform. For those who have used Jupyter Notebooks before, this will be a good refresher, and you are likely to uncover new things as well.
What Is a Jupyter Notebook and Why Is It Useful?
Jupyter Notebooks are locally run on web applications that contain live code, equations, figures, interactive apps, and Markdown text in which the default programming language is Python. In other words, a Notebook will assume you are writing Python unless you tell it otherwise. We'll see examples of this when we work through our first workbook, later in this chapter.
Note
Jupyter Notebooks support many programming languages through the use of kernels, which act as bridges between the Notebook and the language. These include R, C++, and JavaScript, among many others. A list of available kernels can be found here: https://packt.live/2Y0jKJ0.
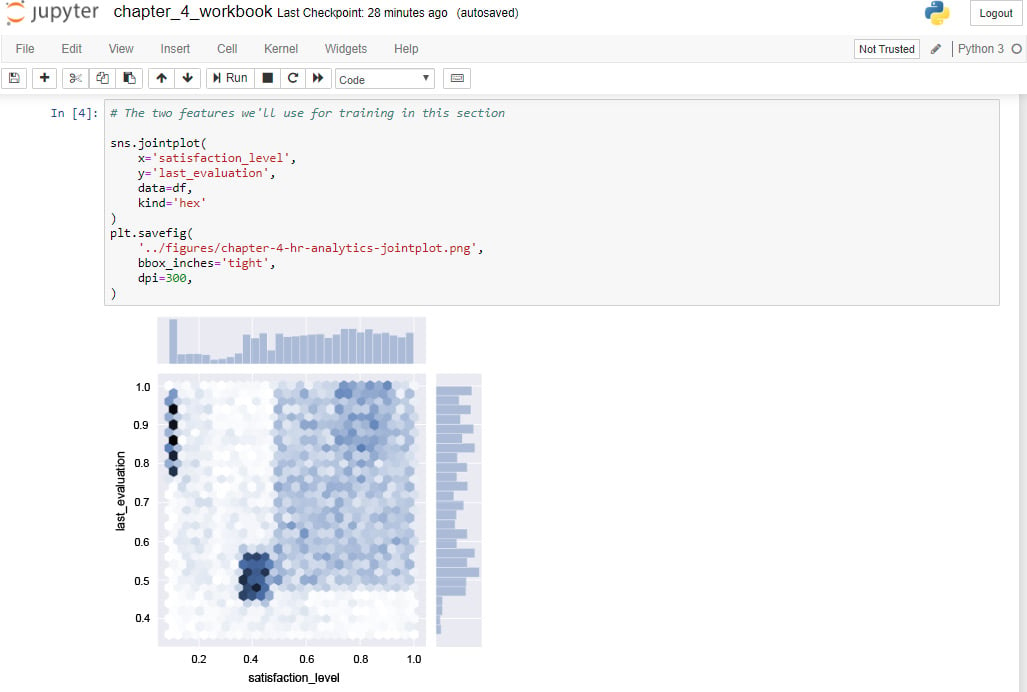
The following is an example of a Jupyter Notebook:
Figure 1.1: Jupyter Notebook sample workbook
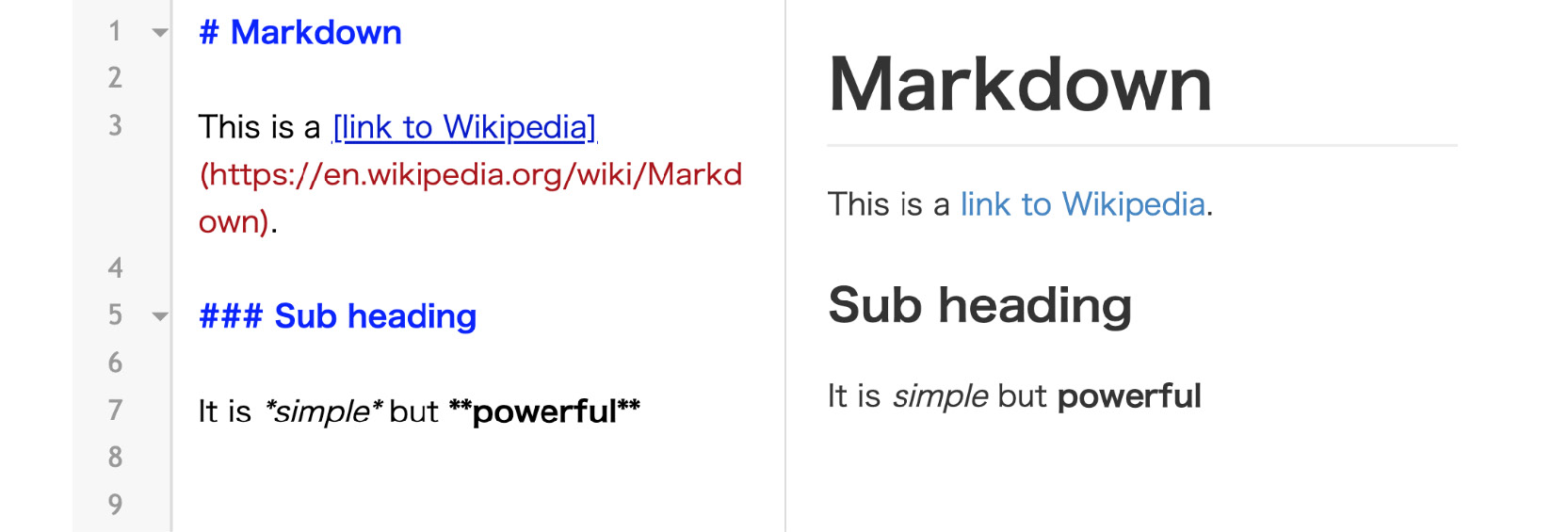
Besides executing Python code, you can write in Markdown to quickly render formatted text, such as titles, lists, or bold font. This can be done in combination with code using the concept of independent cells in the Notebook, as seen in Figure 1.2. Markdown is not specific to Jupyter; it is also a simple language used for styling text and creating basic documents. For example, most GitHub repositories have a README.md file that is written in Markdown format. It's comparable to HTML but offers much less customization in exchange for simplicity.
Commonly used symbols in markdown include hashes (#) to make text into a heading, square ([]) and round brackets (()) to insert hyperlinks, and asterisks (*) to create italicized or bold text:
Figure 1.2: Sample Markdown document
In addition, Markdown can be used to render images and add hyperlinks in your document, both of which are supported in Jupyter Notebooks.
Jupyter Notebooks was not the first tool to use Markdown alongside code. This was the design of R Markdown, a hybrid language where R code can be written and executed inline with Markdown text. Jupyter Notebooks essentially offer the equivalent functionality for Python code. However, as we will see, they function quite differently from R Markdown documents. For example, R Markdown assumes you are writing Markdown unless otherwise specified, whereas Jupyter Notebooks assume you are inputting code. This and other features (as we will explore throughout) make it more appealing to use Jupyter Notebooks for rapid development in data science research.
While Jupyter Notebooks offer a blank canvas for a general range of applications, the types of Notebooks commonly seen in real-world data science can be categorized as either lab-style or deliverable.
Lab-style Notebooks serve as the programming analog of research journals. These should contain all the work you've done to load, process, analyze, and model the data. The idea here is to document everything you've done for future reference. For this reason, it's usually not advisable to delete or alter previous lab-style Notebooks. It's also a good idea to accumulate multiple date-stamped versions of the Notebook as you progress through the analysis, in case you want to look back at previous states.
Deliverable Notebooks are intended to be presentable and should contain only select parts of the lab-style Notebooks. For example, this could be an interesting discovery to share with your colleagues, an in-depth report of your analysis for a manager, or a summary of the key findings for stakeholders.
In either case, an important concept is reproducibility. As long as all the relevant software versions were documented at runtime, anybody receiving a Notebook can rerun it and compute the same results as before. The process of actually running code in a Notebook (as opposed to reading a pre-computed version) brings you much closer to the actual data. For example, you can add cells and ask your own questions regarding the datasets or tweak existing code. You can also experiment with Python to break down and learn about sections of code that you are struggling to understand.
Editing Notebooks with Jupyter Notebooks and JupyterLab
It's finally time for our first exercise. We'll start by exploring the interface of the Jupyter Notebook and the JupyterLab platforms. These are very similar applications for running Jupyter Notebook (.ipynb) files, and you can use whatever platform you prefer for the remainder of this book, or swap back and forth, once you've finished the following exercises.
Note
The .ipynb file extension is standard for Jupyter Notebooks, which was introduced back when they were called IPython Notebooks. These files are human-readable JSON documents that can be opened and modified with any text editor. However, there is usually no reason to open them with any software other than Juptyer Notebook or JupyterLab, as described in this section. Perhaps the one exception to this rule is when doing version control with Git, if you may want to see the changes in plain text.
At this stage, you'll need to make sure that you have the companion material downloaded. This can be downloaded from the open source repository on GitHub at https://packt.live/2zwhfom.
In order to run the code, you should download and install the Anaconda Python distribution for Python 3.7 (or a more recent version). If you already have Python installed and don't want to use Anaconda, you may choose to install the dependencies manually instead (see requirements.txt in the GitHub repository).
Note
Virtual environments are a great tool for managing multiple projects on the same machine. Each virtual environment may contain a different version of Python and external libraries. In addition to Python's built-in virtual environments, conda also offers virtual environments, which tend to integrate better with Jupyter Notebooks.
For the purposes of this book, you do not need to worry about virtual environments. This is because they add complexity that will likely lead to more issues than they aim to solve. Beginners are advised to run global system installs of Python libraries (that is, using the pip commands shown here). However, more experienced Python programmers might wish to create and activate a virtual environment for this project.
We will install additional Python libraries throughout this book, but it's recommended to install some of these (such as mlxtend, watermark, and graphviz) ahead of time if you have access to an internet connection now. This can be done by opening a new Terminal window and running the pip or conda commands, as follows:
graphviz will only be used once, so don't worry too much if you have issues installing it. However, hopefully, you were able to get mlxtend installed since we'll need to rely on it in later chapters to compare models and visualize how they learn patterns in the data.
Exercise 1.01: Introducing Jupyter Notebooks
In this exercise, we'll launch the Jupyter Notebook platform from the Terminal and learn how the visual user interface works. Follow these steps to complete this exercise:
- Navigate to the companion material directory from the Terminal. If you don't have the code downloaded yet, you can clone it using the
git command-line tool:git clone https://github.com/PacktWorkshops/The-Applied-Data-Science-Workshop.git
cd The-Applied-Data-Science-Workshop
Note
With Unix machines such as Mac or Linux, command-line navigation can be done using ls to display directory contents and cd to change directories. On Windows machines, use dir to display directory contents and use cd to change directories. If, for example, you want to change the drive from C: to D:, you should execute D: to change drives. This is an important step if you wish to enable all commands based on folder structure and ensure they run smoothly.
- Run the Jupyter Notebook platform by asking for its version:
Note
The # symbol in the code snippet below denotes a code comment. Comments are added into code to help explain specific bits of logic.
jupyter notebook –-version
# should return 6.0.2 or a similar / more recent version
- Start a new local Notebook server here by typing the following into the Terminal:
jupyter notebook
A new window or tab of your default browser will open the Notebook Dashboard to the working directory. Here, you will see a list of folders and files contained therein.
- Reopen the Terminal window that you used to launch the app. We will see the NotebookApp being run on a local server. In particular, you should see a line like this in the Terminal:
[I 20:03:01.045 NotebookApp] The Jupyter Notebook is running at: http:// localhost:8888/?token=e915bb06866f19ce462d959a9193a94c7 c088e81765f9d8a
Going to the highlighted HTTP address will load the app in your browser window, as was done automatically when starting the app.
- Reopen the web browser and play around with the Jupyter Dashboard platform by clicking on a folder (such as
chapter-06), and then clicking on an .ipynb file (such as chapter_6_workbook.ipynb) to open it. This will cause the Notebook to open in a new tab on your browser.
- Go back to the tab on your browser that contains the Jupyter Dashboard. Then, go back to the root directory by clicking the
… button (above the folder content listing) or the folder icon above that (in the current directory breadcrumb).
- Although its main use is for editing Notebook files, Jupyter is a basic text editor as well. To see this, click on the
requirements.txt text file. Similar to the Notebook file, it will open in a new tab of your browser.
- Now, you need to close the platform. Reopen the Terminal window you used to launch the app and stop the process by typing Ctrl + C in the Terminal. You may also have to confirm this by entering
y and pressing Enter. After doing this, close the web browser window as well.
- Now, you are going to explore the Jupyter Notebook command-line interface (CLI) a bit. Load the list of available options by running the following command:
jupyter notebook --help
- One option is to specify the port for the application to run on. Open the NotebookApp at local port
9000 by running the following command:jupyter notebook --port 9000
- Click
New in the upper right-hand corner of the Jupyter Dashboard and select a kernel from the drop-down menu (that is, select something in the Notebooks section):Figure 1.3: Selecting a kernel from the drop-down menu
This is the primary method of creating a new Jupyter Notebook.
Kernels provide programming language support for the Notebook. If you have installed Python with Anaconda, that version should be the default kernel. Virtual environments that have been properly configured will also be available here.
- With the newly created blank Notebook, click the top cell and type
print('hello world'), or any other code snippet that writes to the screen.
- Click the cell and press Shift + Enter or select
Run Cell from the Cell menu.Any stdout or stderr output from the code will be displayed beneath as the cell runs. Furthermore, the string representation of the object written in the final line will be displayed as well. This is very handy, especially for displaying tables, but sometimes, we don't want the final object to be displayed. In such cases, a semicolon (;) can be added to the end of the line to suppress the display. New cells expect and run code input by default; however, they can be changed to render markdown instead.
- Click an empty cell and change it to accept the Markdown-formatted text. This can be done from the drop-down menu icon in the toolbar or by selecting
Markdown from the Cell menu. Write some text in here (any text will do), making sure to utilize Markdown formatting symbols such as #, and then run the cell using Shift + Enter:Figure 1.4: Menu options for converting cells into code/Markdown
- Scroll to the
Run button in the toolbar:Figure 1.5: Toolbar icon to start cell execution
- This can be used to run cells. As you will see later, however, it's handier to Shift + Enter to run cells.
- Right next to the Run button is a
Stop icon, which can be used to stop cells from running. This is useful, for example, if a cell is taking too long to run:Figure 1.6: Toolbar icon to stop cell execution
- New cells can be manually added from the
Insert menu:Figure 1.7: Menu options for adding new cells
- Cells can be copied, pasted, and deleted using icons or by selecting options from the
Edit menu:Figure 1.8: Toolbar icons to cut, copy, and paste cells
The drop-down list from the Edit menu is as follows:
Figure 1.9: Menu options to cut, copy, and paste cells
- Cells can also be moved up and down this way:
Figure 1.10: Toolbar icons for moving cells up or down
There are useful options in the Cell menu that you can use to run a group of cells or the entire Notebook:
Figure 1.11: Menu options for running cells in bulk
Experiment with the toolbar options to move cells up and down, insert new cells, and delete cells. An important thing to understand about these Notebooks is the shared memory between cells. It's quite simple; every cell that exists on the sheet has access to the global set of variables. So, for example, a function defined in one cell could be called from any other, and the same applies to variables. As you would expect, anything within the scope of a function will not be a global variable and can only be accessed from within that specific function.
- Open the
Kernel menu to see the selections. The Kernel menu is useful for stopping the execution of the script and restarting the Notebook if the kernel dies: Figure 1.12: Menu options for selecting a Notebook kernel
Kernels can also be swapped here at any time, but it is inadvisable to use multiple kernels for a single Notebook due to reproducibility concerns.
- Open the
File menu to see the selections. The File menu contains options for downloading the Notebook in various formats. It's recommended to save an HTML version of your Notebook, where the content is rendered statically and can be opened and viewed as you would expect in web browsers.
- The Notebook name will be displayed in the upper left-hand corner. New Notebooks will automatically be named
Untitled. You can change the name of your .ipynb Notebook file by clicking on the current name in the upper left corner and typing in the new name. Then, save the file.
- Close the current tab in your web browser (exiting the Notebook) and go to the Jupyter Dashboard tab, which should still be open. If it's not open, then reload it by copying and pasting the HTTP link from the Terminal.
- Since you didn't shut down the Notebook (you just saved and exited it), it will have a green book symbol next to its name in the
Files section of the Jupyter Dashboard, and it will be listed as Running on the right-hand side next to the last modified date. Notebooks can be shut down from here.
- Quit the Notebook you have been working on by selecting it (checkbox to the left of the name) and clicking the orange
Shutdown button.Note
If you plan to spend a lot of time working with Jupyter Notebooks, it's worthwhile learning the keyboard shortcuts. This will speed up your workflow considerably. Particularly useful commands to learn are the shortcuts for manually adding new cells and converting cells from code into Markdown formatting. Click on Keyboard Shortcuts from the Help menu to see how.
- Go back to the Terminal window that's running the Jupyter Notebook server and shut it down by typing Ctrl + C. Confirm this operation by typing
y and pressing Enter. This will automatically exit any kernel that is running. Do this now and close the browser window as well.
Now that we have learned the basics of Jupyter Notebooks, we will launch and explore the JupyterLab platform.
While the Jupyter Notebook platform is lightweight and simple by design, JupyterLab is closer to R Studio in design. In JupyterLab, you can stack notebooks side by side, along with console environments (REPLs) and data tables, among other things you may want to look at.
Although the new features it provides are nice, the simplicity of the Jupyter Notebook interface means that it's still an appealing choice. Aside from its simplicity, you may find the Jupyter Notebook platform preferable for the following reasons:
- You may notice minor latency issues in JupyterLab that are not present in the Jupyter Notebook platform.
- JupyterLab can be extremely slow to load large
.ipynb files (this is an open issue on GitHub, as of early 2020).
Please don't let these small issues hold you back from trying out JupyterLab. In fact, it would not be surprising if you decide to use it for running the remainder of the exercises and activities in this book.
The future of open source tooling around Python and data science is going to be very exciting, and there are sure to be plenty of developments regarding Jupyter tools in the years to come. This is all thanks to the open source programmers who build and maintain these projects and the companies that contribute to the community.
Exercise 1.02: Introducing the JupyterLab Platform
In this exercise, we'll launch the JupyterLab platform and see how it compares with the Jupyter Notebook platform.
Follow these steps to complete this exercise:
- Run JupyterLab by asking for its version:
jupyter lab --version
# should return 1.2.3 or a similar / more recent version
- Navigate to the root directory, and then, launch JupyterLab by typing the following into the Terminal:
jupyter lab
Similar to when we ran the Jupyter Notebook server, a new window or tab on your default browser should open the JupyterLab Dashboard. Here, you will see a list of folders and files in the working directory in a navigation bar to the left:
Figure 1.13: JupyterLab dashboard
- Looking back at the Terminal, you can see a very similar output to what our NotebookApp showed us before, except now for the LabApp. If nothing else is running there, it should launch on port
8888 by default:[I 18:37:29.369 LabApp] The Jupyter Notebook is running at:
[I 18:37:29.369 LabApp] http://localhost:8888/?token=cb55c8f3c03f0d6843ae59e70bedbf3b6ec 4a92288e65fa3
- Looking back at the browser window, you can see that the JupyterLab Dashboard has many of the same menus as the Jupyter Notebook platform. Open a new Notebook by clicking
File | New | Notebook:Figure 1.14: Opening a new notebook
- When prompted to select a kernel, choose
Python 3:Figure 1.15: Selecting a kernel for our notebook
The Notebook will then load into a new tab inside JupyterLab. Notice how this is different from the Jupyter Notebook platform, where each file is opened in its own browser tab.
- You will see that a toolbar has appeared at the top of the tab, with the buttons we previously explored, such as those to save, run, and stop code:
Figure 1.16: JupyterLab toolbar and Notebook tab
- Run the following code in the first cell of the Notebook to produce some output in the space below by Shift + Enter:
for i in range(10):
print(i, i % 3)
This will look as follows in Jupyter Notebook:
Figure 1.17: Output of the for loop
- When you place your mouse pointer in the white space present to the left of the cell, you will see two blue bars appear to the left of the cell. This is one of JupyterLab's new features. Click on them to hide the code cell or its output:
Figure 1.18: Bars that hide/show cells and output in JupyterLab
- Explore window stacking in JupyterLab. First, save your new Notebook file by clicking
File | Save Notebook As and giving it the name test.ipynb:Figure 1.19: Prompt for saving the name of the file
- Click
File | New | Console in order to load up a Python interpreter session:Figure 1.20: Opening a new console session
- This time, when you see the kernel prompt, select
test.ipynb under Use Kernel from Other Session. This feature of JupyterLab allows each process to have shared access to variables in memory:Figure 1.21: Electing the console kernel
- Click on the new
Console window tab and drag it down to the bottom half of the screen in order to stack it underneath the Notebook. Now, define something in the console session, such as the following:a = 'apple'
It will look as follows:
Figure 1.22: Split view of the Notebook and console in JupyterLab
- Run this cell with Shift + Enter (or using the
Run menu), and then run another cell below to test that your variable returns the value as expected; for example, print(a).
- Since you are using a shared kernel between this console and the Notebook, click into a new cell in the
test.ipynb Notebook and print the variable there. Test that this works as expected; for example, print(a):Figure 1.23: Sharing a kernel between processes in JupyterLab
A great feature of JupyterLab is that you can open up and work on multiple views of the same Notebook concurrently—something that cannot be done with the Jupyter Notebook platform. This can be very useful when working in large Notebooks where you want to frequently look at different sections.
- You can work on multiple views of
test.ipynb by right-clicking on its tab and selecting New View for Notebook: Figure 1.24: Opening a new view for an open Notebook
You should see a copy of the Notebook open to the right. Now, start typing something into one of the cells and watch the other view update as well:
Figure 1.25: Two live views of the same Notebook in JupyterLab
There are plenty of other neat features in JupyterLab that you can discover and play with. For now, though, we are going to close down the platform.
- Click the circular button with a box in the middle on the far left-hand side of the Dashboard. This will pull up a panel showing the kernel sessions open right now. You can click
SHUT DOWN to close anything that is open:Figure 1.26: Shutting down Notebook sessions in JupyterLab
- Go back to the Terminal window that's running the JupyterLab server and shut it down by pressing Ctrl + C, then confirm the operation by pressing
Y and pressing Enter. This will automatically exit any kernel that is running. Do this now and close the browser window as well:
Figure 1.27: Shutting down the LabApp
In this exercise, we learned about the JupyterLab platform and how it compares to the older Jupyter Notebook platform for running Notebooks. In addition to learning about the basics of using the app, we explored some of its awesome features, all of which can help your data science workflow.
In the next section, we'll learn about some of the more general features of Jupyter that apply to both platforms.