Using IterativeImputer() from scikit-learn, we can model variables using multiple algorithms, such as Bayes, k-nearest neighbors, decision trees, and random forests. Perform the following steps to do so:
- Import the required Python libraries and classes:
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import BayesianRidge
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import ExtraTreesRegressor
from sklearn.neighbors import KNeighborsRegressor
- Load the data and separate it into train and test sets:
variables = ['A2','A3','A8', 'A11', 'A14', 'A15', 'A16']
data = pd.read_csv('creditApprovalUCI.csv', usecols=variables)
X_train, X_test, y_train, y_test = train_test_split(
data.drop('A16', axis=1), data['A16'], test_size=0.3,
random_state=0)
- Build MICE imputers using different modeling strategies:
imputer_bayes = IterativeImputer(
estimator=BayesianRidge(),
max_iter=10,
random_state=0)
imputer_knn = IterativeImputer(
estimator=KNeighborsRegressor(n_neighbors=5),
max_iter=10,
random_state=0)
imputer_nonLin = IterativeImputer(
estimator=DecisionTreeRegressor(
max_features='sqrt', random_state=0),
max_iter=10,
random_state=0)
imputer_missForest = IterativeImputer(
estimator=ExtraTreesRegressor(
n_estimators=10, random_state=0),
max_iter=10,
random_state=0)
Note how, in the preceding code block, we create four different MICE imputers, each with a different machine learning algorithm which will be used to model every variable based on the remaining variables in the dataset.
- Fit the MICE imputers to the train set:
imputer_bayes.fit(X_train)
imputer_knn.fit(X_train)
imputer_nonLin.fit(X_train)
imputer_missForest.fit(X_train)
- Impute missing values in the train set:
X_train_bayes = imputer_bayes.transform(X_train)
X_train_knn = imputer_knn.transform(X_train)
X_train_nonLin = imputer_nonLin.transform(X_train)
X_train_missForest = imputer_missForest.transform(X_train)
Remember that scikit-learn transformers return NumPy arrays.
- Convert the NumPy arrays into dataframes:
predictors = [var for var in variables if var !='A16']
X_train_bayes = pd.DataFrame(X_train_bayes, columns = predictors)
X_train_knn = pd.DataFrame(X_train_knn, columns = predictors)
X_train_nonLin = pd.DataFrame(X_train_nonLin, columns = predictors)
X_train_missForest = pd.DataFrame(X_train_missForest, columns = predictors)
- Plot and compare the results:
fig = plt.figure()
ax = fig.add_subplot(111)
X_train['A3'].plot(kind='kde', ax=ax, color='blue')
X_train_bayes['A3'].plot(kind='kde', ax=ax, color='green')
X_train_knn['A3'].plot(kind='kde', ax=ax, color='red')
X_train_nonLin['A3'].plot(kind='kde', ax=ax, color='black')
X_train_missForest['A3'].plot(kind='kde', ax=ax, color='orange')
# add legends
lines, labels = ax.get_legend_handles_labels()
labels = ['A3 original', 'A3 bayes', 'A3 knn', 'A3 Trees', 'A3 missForest']
ax.legend(lines, labels, loc='best')
plt.show()
The output of the preceding code is as follows:
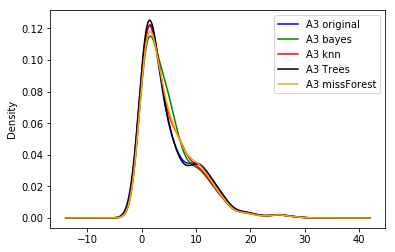
In the preceding plot, we can see that the different algorithms return slightly different distributions of the original variable.











































































